GSCA
- Gene Set Control Analysis
Case Study
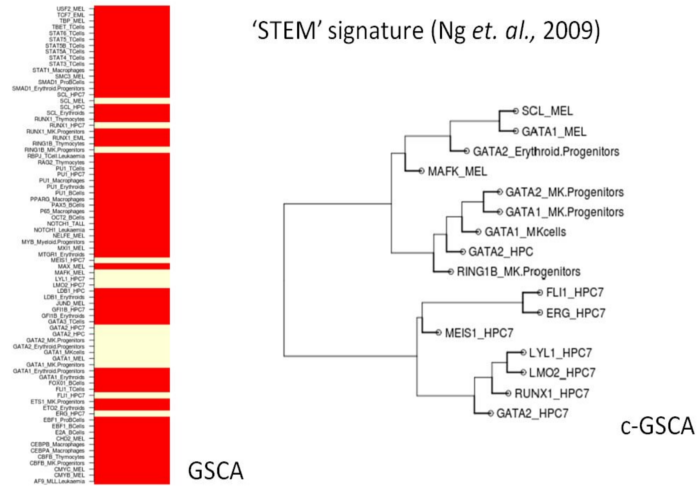
A brief explanation of the functionality of the GSCA web tool is provided below using a recent transcriptome analysis of HSCs and early multi-, bi- and unipotent progenitors (Ng. et. al., 2009) which reported 9 gene expression signatures ranging from those characteristic for the most immature HSCs to those affiliated with differentiation into the individual haematopoietic lineages. We interrogated these 9 experimentally obtained gene expression signatures using the GSCA web tool, thus providing an independent test case to examine the biological relevance of predicted combinatorial regulatory signatures in addition to testing the functionality of the web tool.
Users can paste a query gene list or upload it from a file (human or mouse). Upon choosing 'GSCA', a gene list of interest is interrogated against 78 ChIP-Seq datasets across 15 blood cell types. GSCA calculates the significance of overlap between each ChIP-Seq dataset and the gene set of interest and displays all ChIP-Seq datasets with those showing enrichment in cream colour. For example, the self renewing signature ('stem' signature from Ng et al (2009)) is provided as a test dataset for the users, and shows statistically significant overlap with multiple transcription factors in HPC7 and progenitors. When the same 'stem' signature gene list is analysed using C-GSCA, the overrepresented ChIP datasets are clustered into 2 distinct cell type specific clusters 'HPC7' and 'MK progenitors' (Figure below). 6 of the 7 transcription factors in the HPC7 cluster overlap with the heptad signature, a binding pattern we have previously shown is overrepresented in the loci of genes specifically expressed in HSPCs and therefore associated with gene sets specifically expressed in HSCs. Similarly, the gene signature associated with the third wave of the myeloid lineage program ('d-my' signatures) from Ng et al. (2009) shows statistically significant overlap with two combinatorial binding events, Cebpα, Cebpβ, Stat1, P65 and Pu.1 in macrophages and Myb in myeloid progenitors. In addition to showing the functionality of the web tool, these results suggest that combinatorial control signatures generated by C-GSCA have the potential to provide insights into combinatorial transcriptional control mechanisms.